MicroRNAs are small regulatory RNA molecules that control gene expression. Do they influence cell-to-cell variability, and can we use them to better diagnose cancer?
MicroRNAs were first discovered as small regulatory molecules that control developmental timing events in the nematode worm Caenorhabditis elegans. We have since come to appreciate that many mammalian microRNAs can bind hundreds of different gene products in the cell and regulate their output. This makes microRNAs an ideal candidate for understanding why individual cells behave the way they do, and how that behaviour can vary across a population. In fact, microRNAs may be one of the most sensitive and specific indicators of whether a cell is going to behave as benign or malignant, particularly in liquid cancers such as leukemia. We are building a diagnostic platform that can sequence microRNAs from large numbers of single cells using a novel microfluidic device that can enrich for desired molecular subclasses. This would allow much earlier detection of therapy resistant persister cells, and may also give us a clue as to why they enter into this resistant state in the first place. We collaborate with KI faculty Daniel Anderson and Phillip Sharp on this project.
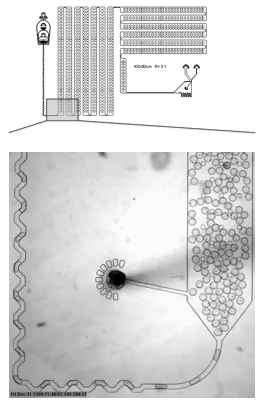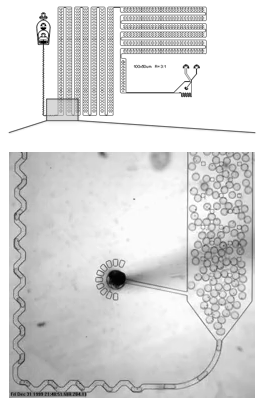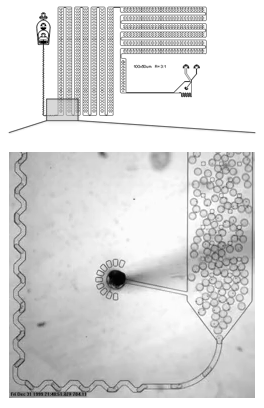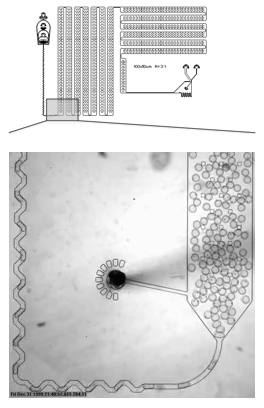
A microfluidic device that encapsulates each cell into a droplet. We have designed holding chambers that allow for enrichment within each droplet of target biomolecules. Our goal is to capture the analytes that best allow us to distinguish normal cells from cancer with the highest precision.

